At its core cnTrack provides a secure and scalable way to analyze, store, retrieve and manage aCGH and SNP array data and soon will become available for Next Generation Seqiencing (NGS) users as well. It was designed to facilitate and monitor distributed data interpretation process involving multiple workstations in a large diagnostic or research laboratory. Using cnTrack laboratory team can standardize and control their work across all major aCGH and SNP array platforms including Affymetrix, Agilent, Illumina, PerkinElmer, Roche-Nimblegen, BlueGnome, OGT and custom arrays. Seamless integration of our analytical and visualization software oneClickCGH and CGH Fusion into cnTrack ensures precise and reliable DNA copy number and LOH analysis in a high-throughput environment.
Features
Streamlined data interpretation and reporting
cnTrack is designed to accelerate DNA Copy Number reporting/interpretation in a modern Cytogenetic laboratory. It easily adapts to any existing array-based platform and any workflow a laboratory may have in place. By integrating all necessary components of Cytogenetic analysis such as clinical data (including connection to Shire, OMNILAB, STARLIMS etc), anomaly detection, data visualization, case reporting and sample management cnTrack has been proven to significantly reduce per-sample processing time and increase throughput.
Well-defined yet scalable reporting pipeline
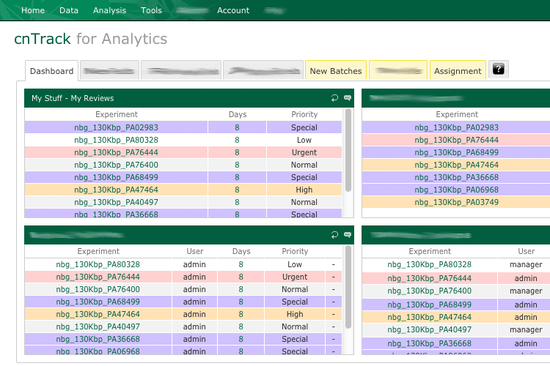
The centerpiece of cnTrack web-based interface is a snapshot of laboratory's reporting pipeline. It can be customized for any particular user role to make distributed data analysis and interpretation more efficient. This snapshot feature ensures that all lab members see their immediate tasks and are aware of any urgent cases. Transparency of cnTrack interface facilitates timely detection and management of any possible bottlenecks in the workflow. cnTrack can be set up for use just by a few users or by dozens across the lab with the number of users adjusted as needed.
Platform-agnostic centralized data management
We built cnTrack to be truly platform agnostic: it accepts array data generated by all major commercial array platforms and can be used on any computer or device that has an Internet browser. Users can store and analyze data produced by major array platforms such as Affymetrix, Agilent, Roche-Nimblegen, Illumina, OGT etc. Import of sequencing data is also available. cnTrack is also agnostic with respect to database technology (Oracle, SQL Server etc) used on its backend for better integration with existing institution infrastructure.
Full spectrum of data analytics
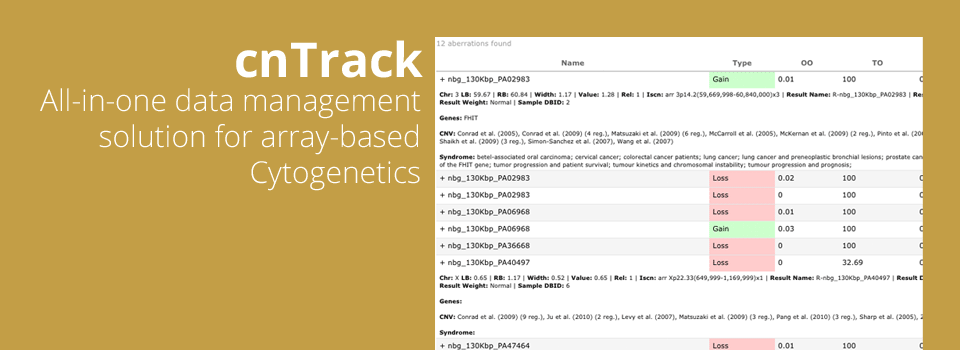
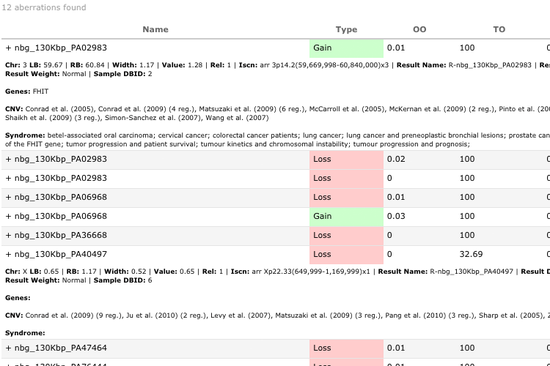
Being integrated with our powerful DNA Copy Number analytics software CGH Fusion, cnTrack delivers full power of Copy Number and LOH analysis to every licensed PC. Data from dozens or even hundreds of samples can be analyzed for chromosomal changes within minutes and then taken through clinical reporting pipeline.
Ease of lab-wide distribution and access
Based on a web-based technology cnTrack does not have any significant performance requirements with respect to user hardware. Cross-laboratory installation effort is also very minimal as most of system’s data management capabilities are delivered via Internet browser. Such light-client approach do data management aspect of cnTrack provides its users with a wide spectrum of infrastructural options. All elements of cnTrack can be installed on a secure server inside institution’s firewall. Alternatively, to avoid server maintenance costs, our clients can choose to keep cnTrack’s database core on a secure cloud managed by
Screenshots
Personal data management space
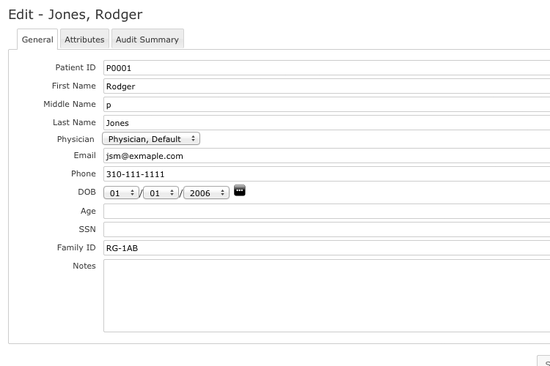
Patient Information
Querying legacy data
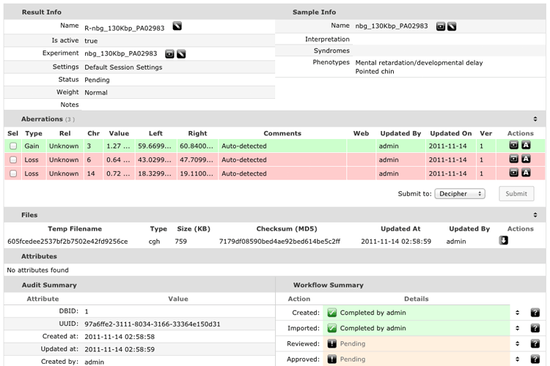
Integrated Cytogenetic report
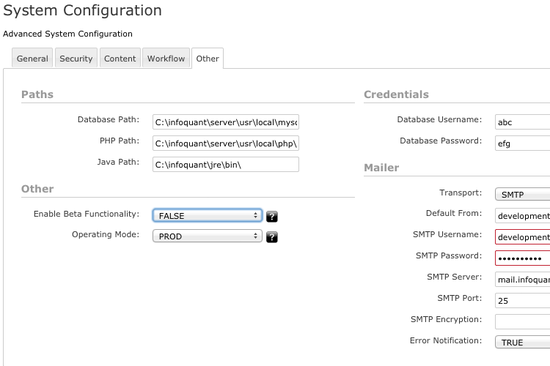
Enterprise-wide Administration
System Requirements
Client
- Windows XP +, OS X 10.6+ , Linux with Java 1.6+
- 2GHz or greater CPU
- 2GB or greater RAM
- 1024x768 or higher display
Server
- Windows XP+, OS X 10.6+ , Linux with Java 1.6+
- Supported RDBMS (SQL Server, Postgres, MySQL, Oracle and others)
- 2GHz or greater CPU
- 4GB or greater RAM
Additional Resources
White Papers
Online Demonstration
infoQuant provides short online demonstrations of cnTrack upon request. Please complete our evaluation request form and a member of our team will contact you to schedule the online meeting.